runchimera.csh
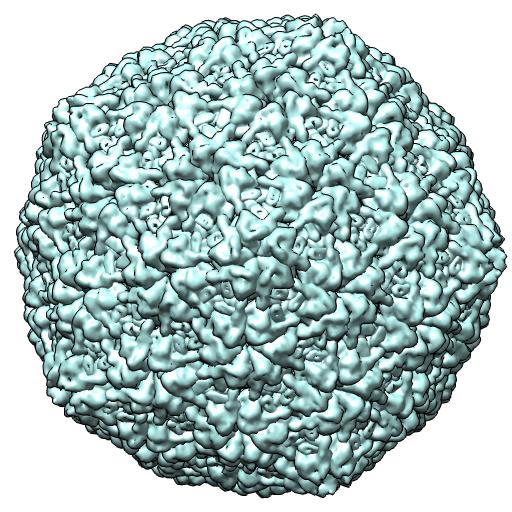
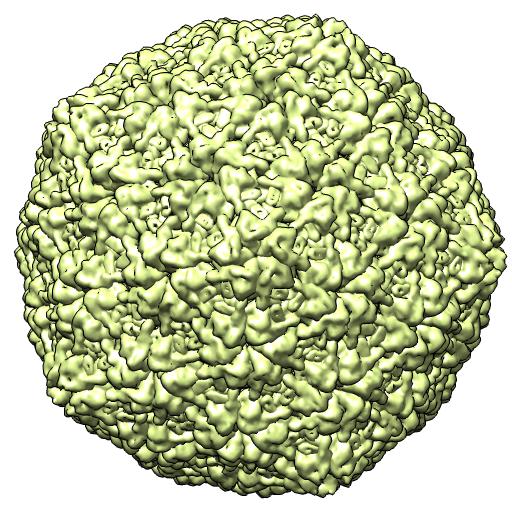
(A) 2XD8 -- EM virus
runpt.csh 2xd8.cif 2xd8.biomt
runchimera.csh
runpt.log assembly.cif
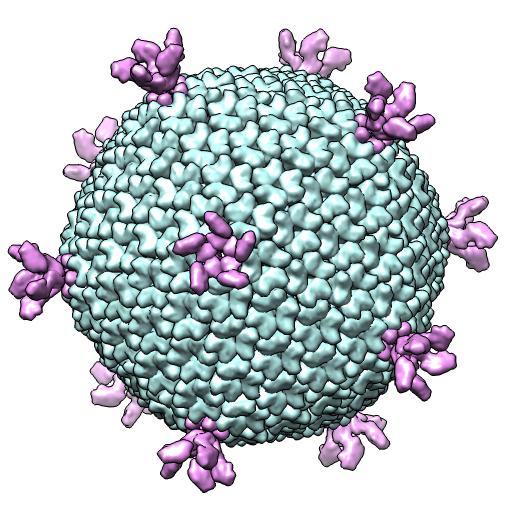
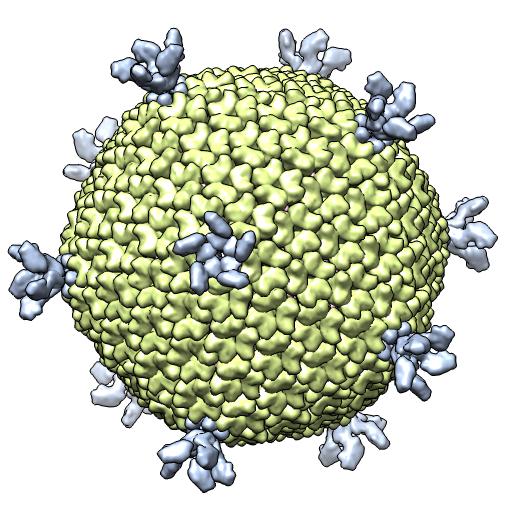
(B) 2W0C -- X-ray 1 particle/crystal au
coordinates in crystal frame (usually true)
runpt.csh 2w0c.cif 2w0c.biomt
runchimera.csh
runpt.log assembly.cif
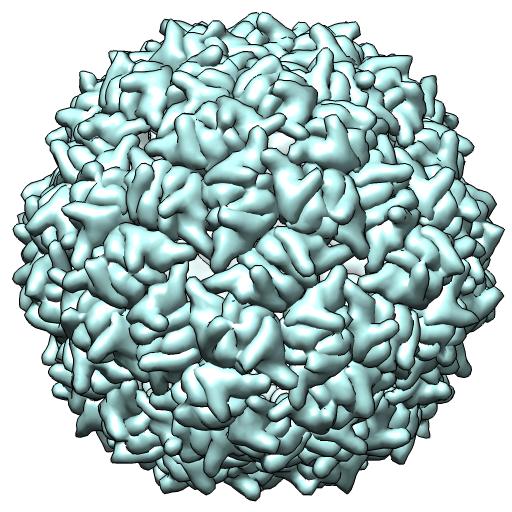
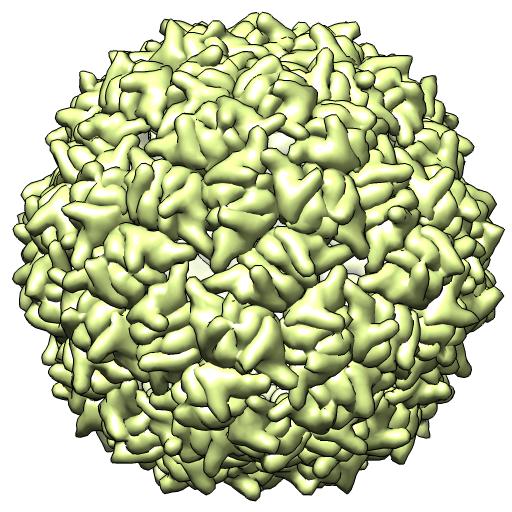
(C) 2VF9 -- X-ray 2 particles/crystal au
coordinates in crystal frame
runpt.csh 2vf9.cif 2vf9.biomt1* ident x1.mat**
*(1st 60 BIOMT matrices in current public file)
**(61st BIOMT matrix in current public file)
runchimera.csh
runpt.log assembly.cif
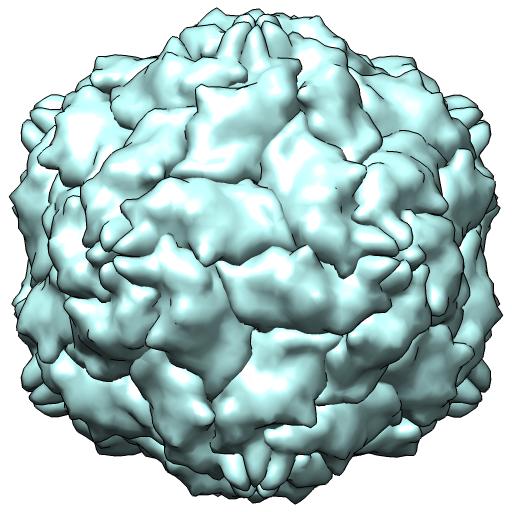
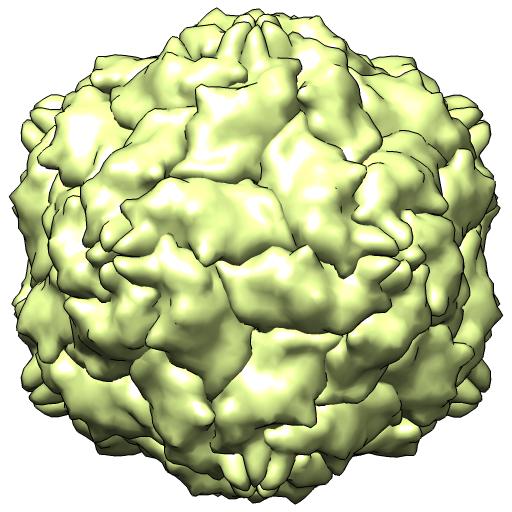
(D) 3N7X -- X-ray 1/2 particle/crystal au
coordinates in NONCRYSTAL frame
runpt.csh 3n7x.cif 3n7x.biomt x0.mat
runchimera.csh
runpt.log assembly.cif